H-Evolution-002
Molecular Biology
Preface
Interest in how nature
replicates began when working with Dr Ken O’Kief of TRW. TRW was tech advisor to AF for the Minuteman
missile program, when I worked for Autonetics division of Rockwell, who built
Guidance and Control systems for that series.
Our work on Post Boost Rocket Engine System had ended and I was out of a
job. I suggested we should determine
how to convert analog controls to digital, and was asked to pursue the
concept. With great difficulty I
developed a digital signal processor that could serve all stages by being programmable. Our companies arranged for Ken and I to work
together on pending concepts. When we
broke for lunch Ken said this is not a signal processor but a programmable
computer, setting the stage for his
question when we returned: “could you
design a computer that could replicate itself?” My thoughts focused on how to automate making the “digital
computer” we’d been discussing. After a
few minutes thought I said, “no, there is no way to automate supply of
parts.” The with a slight smile Ken
said “I’m looking at one.” It took a
moment to realized what he had just said.
We humans are computers that
replicate!!. I had been taking a
night class in Biology and at lunch told Ken of a booklet describing the DNA
molecule and text book “Molecular Biology” by Watson, which prompted his question.
I kept pondering how nature solves the supply problem? When observing contaminate moving in a pool,
I said, that’s it. Life began in the
oceans, a sea of molecular soup, kept in constant motion. The DNA code that evolved was immersed in
it’s part supply, combining parts by magnetic attraction per patterns that
survived.
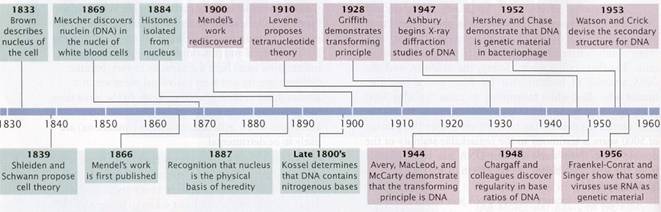
Many contributed to our understanding of genetics
Protein Structure: When a cell makes a polypeptide, the chain
usually folds spontaneously to form the functional shape for that protein (a
biological polymer constructed from amino acid monomers.). It is a protein's
three-dimensional shape that enables the molecule to carry out its specific
function in a cell. In almost every case, a protein's function depends on its
ability to recognize and bind to some other molecule. For example, the
receptors on the brain cell below are actually proteins that recognize certain
chemical signals from other cells. If the protein receptor's shape were to be
altered, then it would not be able to perform this recognition function. With
proteins, function follows form -- that is, what a protein does is a
consequence of its shape.
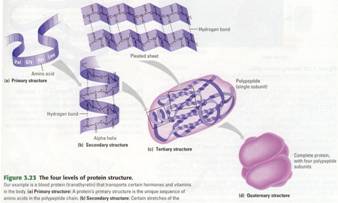
Proteins come in a variety of kinds and
packages, they are the supply source when building cells from DNA & RNA
patterns.
A
protein's shape is sensitive to the surrounding environment. An unfavorable
change in temperature, pH, or some other quality of the aqueous environment can
cause a protein to unravel and lose its normal shape. This is called
denaturation of the protein.
Given an
environment suitable for that protein (so that it doesn't denature), the
primary structure of a protein causes it to fold into its functional shape.
Each kind of protein has a unique primary structure and therefore a unique
shape that enables it to do a certain job in a cell. Each polypeptide chain has a sequence specified by an inherited
gene.
Nucleic
Acids
Nucleic acids
are information storage molecules that provide the directions for building
proteins. The name nucleic comes from their location in the nuclei of
eukaryotic cells. There are actually two types of nucleic acids: DNA (those
most famous of chemical initials, which stand for deoxyribonucleic acid) and
RNA (for ribonucleic acid). The genetic material that humans and other
organisms inherit from their parents consists of giant molecules of DNA. Within
the DNA are genes, specific stretches of DNA that program the amino acid
sequences (primary structure) of proteins. Those programmed instructions,
however, are written in a kind of chemical code that must be translated
from "nucleic acid language" to "protein language."
A cell's RNA molecules help translate.


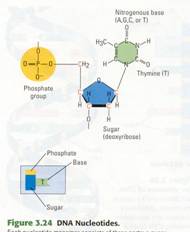
Nucleic acids are
polymers of monomers called nucleotides (Figure 3.24). Each nucleotide is
itself a complex organic molecule with three parts. At the center of each
nucleotide is a five-carbon sugar, deoxyribose in DNA and ribose in RNA. Attached
to the sugar is a negatively charged phosphate group containing a phosphorus
atom bonded to oxygen atoms (PO4-). Also attached to the sugar is a
nitrogen-containing base (nitrogenous base) made of one or two rings. (It is
called a base because it behaves like a base, or alkali, in aqueous solutions.)
The sugar and phosphate are the same in all nucleotides; only the base varies.
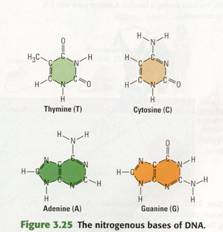
Each DNA nucleotide has one of the following four bases: adenine (abbreviated
A), guanine (G), cytosine (C), or thymine (T). Thus, all genetic information is
written in a four-letter alphabet-A, G, C, T -the bases that distinguish the
four nucleotides that make up DNA
(Figure 3.25).
Nucleotide monomers are linked into long
chains called polynucleotides, or DNA strands (Figure 3.26a). Nucleotides are
joined together by covalent .bonds between the sugar of one nucleotide and the
phosphate of the next. This results in a sugar-phosphate backbone, a repeating
pattern of sugar-phosphate-sugar-phosphate, with the bases hanging off the backbone
like appendages. Polynucleotides vary in length from long to very long, so the
number of possible polynucleotide sequences is very great. One long DNA strand
contains many genes, each a specific series of hundreds or thousands of
nucleotides. And each of these genes stores information in its unique sequence
of nucleotide bases. In fact, it is this information that cells translate into
an amino acid sequence to make a specific protein.
How can the cells of parents copy their
genes to pass along to offspring? Inheritance is based on DNA actually being
double-stranded, with the two DNA strands wrapped around each other to form a
double helix. In the central core of the helix, the bases along one DNA strand
hydrogen-bond to bases along the other strand. This base pairing is specific:
The base A can pair only with T, and G can pair only with C. Thus, if you know
the sequence of bases along one DNA strand, you also know the sequence along
the complementary strand in the double helix. This unique base pairing is the
basis of DNA’s ability to act as the molecule of inheritance.
What about RNA? As its name-ribonucleic
acid-implies, its sugar is ribose rather than deoxyribose. By comparing the RNA
nucleotide in, Figure 3.27 with the DNA nucleotide in Figure 3.24 you can see
that the RNA ribose sugar has an extra -OH group compared with the DNA
deoxyribose sugar (deoxy means "without an oxygen"). Another
difference between RNA and DNA is that instead of the base thymine, RNA has a
similar but distinct base called uracil (U). Except for the presence of ribose
and uracil, an RNA polynucleotide chain is identical to a DNA polynucleotide
chain. However, RNA is usually found only in a single-stranded form, while
DNA usually exists as a double helix.

Figure
10-2
Figure 10.2 The structure of a DNA
polynucleotide. A molecule of DNA contains two polynucleotides, each a chain of
nucleotides. Each nucleotide consists of a nitrogenous base, a sugar (blue),
and a phosphate group (gold). The nucleotides are linked, the sugar of one
connected to the phosphate of the next, forming a sugar-phosphate backbone,
with the bases protruding from the sugars. The chemical structure at the right
shows the details of a DNA nucleotide. The sugar has five carbon atoms (shown
in red for emphasis) and is called deoxyribose. The phosphate group has given
up an H+ ion, acting as an acid. Hence, the full name for DNA is
deoxyribonucleic acid.
The Structure and Replication of DNA
DNA was known as a chemical in cells by
the end of the nineteenth century, but Mendel and other early geneticists did
all their work without any know-ledge of DNA’s role in heredity. By the late
1930s, experimental studies had convinced most biologists that a specific kind
of molecule, rather than some complex chemical mixture, was the basis of
inheritance. Attention focused on
chromosomes, which were already known to carry genes. By the 1940s, scientists
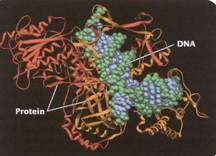
knew that chromosomes consisted of two types of chemicals: DNA and protein. And
by the early 1950s, a series of discoveries had convinced the scientific world
that DNA was the hereditary material.
What came next was one of the most
celebrated quests in the history of science -- the effort to figure out the
structure of DNA. A good deal was already known about DNA. Scientists had
identified all its atoms and knew how they were covalently bonded to one
another. What was not understood was the specific arrangement that gave DNA its
unique properties – the capacity to store genetic information, copy it, and
pass it - from generation to generation. A race was on to discover how the
structure of this molecule could account for its role in heredity. We will
describe that momentous discovery shortly. First, look at the underlying
chemical structure of DNA and its chemical cousin RNA.
DNA
and RNA: Polymers of Nucleotides I
Both DNA and RNA are nucleic acids, which
consist of long chains (polymers) of chemical units (monomers) called
nucleotides. (Figures 3.25-3.27.) A very simple diagram of a nucleotide
polymer, or polynucleotide, is shown on the left in Figure 10.2. This sample
polynucleotide chain shows only one possible arrangement of the four different
types of nucleotides ( abbreviated A, C, T and G) that make up DNA.
Polynucleotides tend to be very long and can have any sequence of nucleotides,
so a great number of polynucleotide chains are possible.
The nucleotides are joined to one another
by covalent bonds between the sugar of one nucleotide and the phosphate of the
next. This results in a sugar-phosphate backbone, a repeating pattern of
sugar-phosphate-sugar-phosphate. The nitrogenous bases are arranged like
appendages along this backbone. Zooming in on our polynucleotide in Figure
10.2, we see that each nucleotide consists of three components: a nitrogenous
base, a sugar (blue), and a phosphate group (gold). Examining a single
nucleotide even more closely (Figure 10.2), we see the chemical structure of
its three components. The phosphate group, with a phosphorus atom (P) at its
center, is the source of the acid in nucleic acid. (The phosphate has given up
a hydrogen ion, H+ , leaving a negative charge on one of its oxygen atoms.) The
sugar has five carbon atoms: four in its ring and one extending above the ring.
The ring also includes an oxygen atom. The sugar is called deoxyribose because,
compared to the sugar ribose, it is missing an oxygen atom. The full name for
DNA is deoxyribonucleic acid, with the nucleic part coming from DNA’s location
in the nuclei of eukaryotic cells. (some, mDNA, is located in the
mitochondria.) The nitrogenous base ( thymine, in our example) has a ring of
nitrogen and carbon atoms with various functional groups attached. In contrast
to the acidic phosphate group, nitrogenous bases are basic; hence their name.

Figure 10.3 Co discoverers
Watson & Crick
Figure 10.4 A rope-ladder
model of a double helix. The ropes at the sides represent the sugar-phosphate
backbones. Each wooden rung stands for a pair of bases connected by hydrogen
bonds.
The four nucleotides found in DNA differ
only in their nitrogenous bases (Figure 3.25). At this point, the structural
details are not as important as the fact that the bases are of two types. Thymine
(T) and cytosine (C) are single-ring structures. Adenme (A)
and guanme (G) are larger, double-ring structures. (The one-letter
abbreviations can be used for either the bases alone or for the nucleotides
containing them.) Recall that RNA has the nitrogenous base uracil (U)
instead of thymine ( uracil is very similar to thymine). RNA also contains a
slightly different sugar than DNA (ribose instead of deoxyribose). Other than
that, RNA and DNA polynucleotides have the same chemical structure.
Watson
and Crick's Discovery of the Double Helix
The celebrated partnership that resulted
in the determination of the physical structure of DNA began soon after a
23-year-old American named James D. Watson journeyed to Cambridge University,
where Englishman Francis Crick was studying protein structure with a technique
called X-ray crystallography . While visiting the laboratory of Maurice Wilkins
at King's College in London, Watson saw an X-ray crystallographic photograph of
DNA, produced by Wilkins's colleague Rosalind Franklin. The photograph clearly
revealed the basic shape of DNA to be a helix (spiral). On the basis of
Watson's later recollection of the photo, he and Crick deduced that the
diameter of the helix was uniform. The thickness of the helix suggested that it
was made up of two polynucleotide strands-in other words, a double helix.
Using wire models, Watson and Crick began
trying to construct a double helix that would conform both to Franklin's data
and to what was then known about the chemistry of DNA. After failing to make a
satisfactory model that placed the sugar-phosphate backbones inside the double
helix, Watson tried putting the backbones on the outside and forcing the
nitrogenous bases to swivel to the interior of the molecule. It occurred to him
that the four kinds of bases might pair in a specific way. This idea of
specific base pairing was a flash of inspiration that enabled Watson and Crick
to solve the DNA puzzle.
At first, Watson imagined that the bases
paired like with like - for example A with A, C with C. But that kind of
pairing did not fit with the fact

Figure
10.5
Figure 10.5 Three representations of DNA. (a) In this model, the
sugar-phosphate backbones are blue ribbons, and the bases are complementary
shapes in shades of green and orange. (b) In this more chemically detailed
structure, you can see the individual hydrogen bonds (dashed lines). You can
also see that the strands run in opposite directions; notice that the sugars on
the two strands are upside down with respect to each other. (c) In this
computer graphic of a DNA double helix, each atom is shown as a sphere,
creating a space-filling model, that the DNA molecule was a uniform diameter.
In AA pair (made of double-ringed bases) would be almost twice as wide as a CC
pair (made of single-ringed bases), causing bulges in the molecule. It soon
became apparent that a double-ringed base on one strand must always be paired
with a single-ringed base on the opposite strand. Moreover, Watson and Crick
realized that the individual structures of the bases dictated the pairings even
more specifically. Each base has chemical side groups that can best form
hydrogen bonds with one appropriate partner ( to review the hydrogen bond, see
Figure 2.11). Adenine can best form hydrogen bonds with thymine, and guanine
with cytosine. In the biologist's shorthand, A pairs with T, and G pairs with
C. A is also said to be "complementary" to T, and G to C.
You can picture the model of the DNA
double helix proposed by Watson and Crick as a rope ladder having rigid, wooden
rungs, with the ladder twisted into a spiral (Figure 10.4). Figure 10.5 shows
three more detailed representations of the double helix. The ribbon like
diagram in Figure 10.5a symbolizes the models of the bases with shapes that
emphasize their complementarity. Figure 10.5b is a more chemically precise
version showing only four base pairs, with the helix untwisted and the
individual hydrogen bonds specified by dashed lines; you can see that the
double helix has an antiparallel arrangement-that is, the two sugar-phosphate
backbones are oriented in opposite directions. Figure 10.5c is a computer
graphic showing every atom of part of a double helix.
Although the base-pairing rules dictate
the side-by-side combinations nitrogenous bases that form the rungs of the
double helix, they place no restrictions on the sequence of nucleotides along
the length of a DNA strand. In fact, the sequence of bases can vary in
countless ways.
In April 1953, Watson and Crick shook the
scientific world with a succinct, two-page announcement of their molecular
model for DNA in the journal Nature. Few milestones in the history of biology
have had as broad an impact as their double helix, with its AT and CG base
pairing. In 1962, Watson, Crick, and Wilkins received the Nobel Prize for their
work. {Franklin may have received the prize as well, had she not died from
cancer in 1958.)
In their 1953 paper, Watson and Crick
wrote that the structure they proposed "immediately suggests a possible
copying mechanism for the genetic material." In other words, the structure
of DNA also points toward a molecular explanation for life's unique properties
of reproduction and inheritance, as we see next.
DNA
Replication
When a cell or a whole organism
reproduces, a complete set of genetic instructions must pass from one
generation to the next. For this to occur, there must be a means of copying the
instructions. Watson and Crick's model for DNA structure immediately suggested
to them that DNA replicates by a template mechanism-each DNA strand can
serve as a mold, or template, to guide reproduction of the other strand.
The logic behind the Watson-Crick proposal for how DNA is copied is quite
simple. If you know the sequence of bases in one strand of the double helix,
you can very easily determine the sequence of bases in the other strand by
applying the base-pairing rules: A pairs with T (and T with A), and G pairs
with C (and C with G). For example, if one polynucleotide has the sequence
ATCG, then the complementary polynucleotide in that DNA molecule must have the
sequence TAGC.
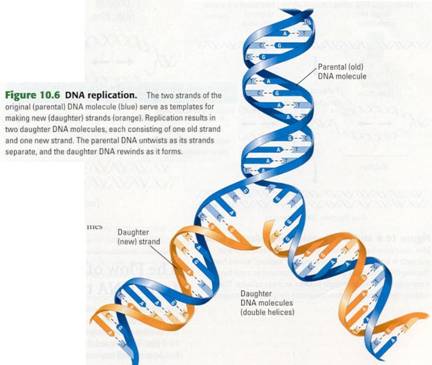
Figure 10.6 shows how the template model
can account for the direct copying of a piece of DNA. The two strands of
parental DNA separate, and each becomes a template for the assembly of a
complementary strand from a supply of free nucleotides. The nucleotides are
lined up one at a time along the template strand in accordance with the
base-pairing rules. Enzymes link the nucleotides to form the new DNA strands.
The completed new molecules, identical to the parental molecule, are known as
daughter DNA molecules (no gender should be inferred from this name).
Although the general mechanism of DNA replication
is conceptually simple, the actual process is complex and requires the
cooperation of more than a dozen enzymes and other proteins. The enzymes that
actually make the covalent bonds between the nucleotides of a new DNA strand
are called DNA polymerases. As an incoming nucleotide base-pairs with
its complement on the template strand, a DNA polymerase adds it to the end of
the growing daughter strand (polymer ). The process is both fast and
amazingly accurate; typically, DNA replication proceeds at a rate of 50
nucleotides per second, with only about one in a billion incorrectly
paired. (This is like parallel signal processing) In addition to their
roles in DNA replication, DNA polymerases and some of the associated proteins
are also involved in repairing damaged DNA. DNA can be harmed by toxic
chemicals in the environment or by high-energy radiation, such as X-rays and
ultraviolet light
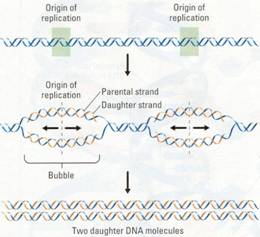

DNA replication begins at specific sites
on a double helix, called origins of replication. It then proceeds in both
directions, creating what are called replication "bubbles" (Figure
10.8). The parental DNA strands open up as daughter strands elongate on both
sides of each bubble. The DNA molecule of a eukaryotic chromosome has many
origins where replication can start simultaneously, shortening the total time
needed for the process. Eventually, all the bubbles merge, yielding two
completed double-stranded daughter DNA molecules.
DNA replication ensures that all the
somatic cells in a multicellular organism carry the same genetic information.
It is also the means by which genetic instructions are copied for the next
generation of the organism.
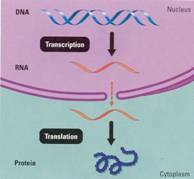

How
an Organism's DNA Genotype Produces Its Phenotype
Knowing the structure of DNA, we can now
define genotype (genetic makeup) and phenotype (the
expressed traits of an organism) more precisely. An organism's genotype is the sequence of nucleotide bases in its
DNA. The molecular basis of the phenotype lies in proteins with a variety of
functions. For example, structural proteins help make up the body of an
organism, and enzymes catalyze its metabolic activities. What is the connection
between the genotype and the protein molecules that more directly determine the
phenotype? Recall that DNA specifies the synthesis of proteins. A gene does
not build a protein directly, but rather dispatches instructions in the form of
RNA, which in turn programs protein synthesis. This central concept in
biology is summarized in Figure 10.9. The chain of command is from DNA in the
nucleus of the cell to RNA to protein synthesis in the cytoplasm (the parts
supply) . The two main stages are transcription, the transfer of genetic
information from DNA into an RNA molecule, and translation, the transfer
of the information in the RNA into a protein.
The relationship between genes and
proteins was first proposed in 1909, when English physician Archibald Garrod
suggested that genes dictate phenotypes through enzymes, the proteins that
catalyze chemical processes. Garrod's idea came from his observations of
inherited diseases. He hypothesized that an inherited disease reflects a
person's inability to make a particular enzyme, and he referred to such
diseases as "inborn errors of metabolism." He gave as one example the
hereditary condition called alkaptonuria, in which the urine appears dark red
because it contains a chemical called alkapton. Garrod reasoned that normal
individuals have an enzyme that breaks down alkapton, whereas alkaptonuric
individuals lack the enzyme. Garrod's hypothesis was ahead of its time, but
research conducted decades later proved him right. In the intervening years,
biochemists accumulated evidence that cells make and break down biologically
important molecules via metabolic pathways, as in the synthesis of an amino
acid or the breakdown of a sugar. Each step in a metabolic pathway is catalyzed
by a specific enzyme. If a person lacks one of the enzymes, the pathway cannot
be completed.
The major breakthrough in demonstrating
the relationship between genes and enzymes came in the 1940s from the work of
American geneticists George Beadle and Edward Tatum with the orange bread mold
Neurospora crassa. Beadle and Tatum studied strains of the mold that were
unable to grow on the usual simple growth medium. Each of these strains turned out to lack an enzyme in a metabolic
pathway that produced some molecule the mold needed such as an amino acid.
Beadle and Tatum also showed that each mutant was defective in a single gene.
Accordingly, they formulated the one gene-one enzyme hypothesis, which states
that the function of an individual gene is to dictate the production of a
specific enzyme.
The one gene-one enzyme hypothesis has
been amply confirmed, but with some important modifications. First it was
extended beyond enzymes to include all types of proteins. For example,
alpha-keratin, the structural protein of your hair, is the product of a gene.
So biologists soon began to think in terms of one gene-one protein. Then it was
discovered that many proteins have two or more different polypeptide chains,
and each polypeptide is specified by
its own gene. Thus, Beadle and Tatum's hypothesis has come to be restated as
one gene-one polypeptide.
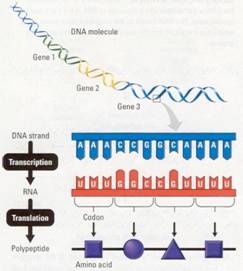


From
Nucleotide Sequence to Amino Acid Sequence: An Overview
Stating that genetic information in DNA is
transcribed into RNA and then translated into polypeptides does not explain how
these processes occur. Transcription and translation are linguistic terms, and
it is useful to think of nucleic acids and polypeptides as having languages,
too. To understand how genetic information passes from genotype to phenotype, we
need to see how the chemical language of DNA is translated into the different
chemical language of polypeptides.
What exactly is the language of nucleic
acids? Both DNA and RNA are polymers made of monomers in specific sequences
that carry information, much as specific sequences of letters carry information
in English. In DNA, the monomers are the four types of nucleotides, which
differ in their nitrogenous bases (A, T, C, and G). The same is true for RNA,
although it has the base U instead of T.
DNAs language is written as a linear
sequence of nucleotide bases, a sequence such as the one you see on the
enlarged DNA strand in Figure 10.10.
Specific sequences of bases, each with a beginning and an end, make up
the genes on a DNA strand. A typical gene consists of thousands of nucleotides,
and a DNA molecule may contain thousands of genes.
When DNA is transcribed, the result is an
RNA molecule. The process is called transcription because the nucleic acid
language of DNA has simply been rewritten (transcribed) as a sequence of bases
of RNA; the language is still that of nucleic acids. The nucleotide bases of
the RNA molecule are complementary to those on the DNA strand. As you will soon
see, this is because the RNA was synthesized using the DNA as a template.
Translation is the conversion of the
nucleic acid language into the polypeptide language. Like nucleic acids,
polypeptides are polymers, but the monomers that make them up -- the letters
of the polypeptide alphabet are the 20 amino acids common to all organisms.
Again, the language is written in a linear sequence, and the sequence of
nucleotides of the RNA molecule dictates the sequence of amino acids of the
polypeptide. But remember, RNA is only a messenger; the genetic information
that dictates the amino acid sequence is based in DNA.
What are the rules for translating the RNA
message into a polypeptide? In other words, what is the correspondence between
the nucleotides of an RNA molecule and the amino acids of a polypeptide? Keep
in mind that there are only four different kinds of nucleotides in DNA (A, G,
C, T) and RNA (A, G, C, U). In translation, these four must somehow specify 20
amino acids. If each nucleotide base coded for one amino acid, only 4 of the 20
amino acids could be accounted for. What if the language consisted of
two-letter code words? If we read the bases of a gene two at a time, AG, for
example, could specify one amino acid, while AT could designate a different
amino acid. However, when the four bases are taken two by two, there are only
16 (that is, 41 possible arrangements-still not enough to specify all 20 amino
acids.
Triplets of bases are
the smallest "words" of uniform length that can specify all the amino
acids. There can be 64 ( that is, 43) possible code words of this
type-more than enough to specify the 20 amino acids. Indeed, there are enough
triplets to allow more than one coding for each amino acid. For example, the
base triplets AAT and AAC could both code for the same amino acid - and, in
fact, they do.
Experiments have verified that the flow of
information from gene to protein is based on a triplet code. The genetic
instructions for the amino acid sequence of a polypeptide chain are written in
DNA and RNA as a series of three-base words called codons. Three-base
codons in the DNA are transcribed into complementary three-base codons in the
RNA, and then the RNA codons are translated into amino acids that form a
polypeptide. Next we turn to the codons themselves.
The
Genetic Code
In 1799, a large stone tablet was found in
Rosetta, Egypt, carrying the same lengthy inscription in three ancient scripts:
Greek, Egyptian hieroglyphics, and Egyptian written in a simplified script. The
Rosetta stone provided the key that enabled scholars to crack the previously
indecipherable hieroglyphic code.
In cracking the genetic code, the set of
rules relating nucleotide sequence to amino acid sequence, scientists wrote
their own Rosetta stone. It was based on a series of elegant experiments , that
revealed the amino acid translations of each of the nucleotide - triplet code
words. The first codon was deciphered in 1961 by American biochemist Marshall
Nirenberg. He synthesized an artificial RNA molecule by linking together
identical RNA nucleotides having uracil as their base. No matter where this
message started or stopped, it could contain only one type of triplet codon:
uuu. Nirenberg added this "poly U" to a test tube mixture containing
ribosomes and the other ingredients required for polypeptide synthesis. This
mixture translated the poly U into a polypeptide containing a single kind of
amino acid, phenylalanine. In this way, Nirenberg learned that the RNA codon
UUU specifies the amino acid phenylalanine (Phe). By variations on this method,
the amino acids specified by all the codons were determined.

Figure 10.11 The dictionary of the genetic code, listed by RNA
codons. The three bases of an RNA codon are designated here as the first,
second, and third bases. Practice using this dictionary by finding the codon
UGG. This is the only codon for the amino acid tryptophan (Trp), but most amino
acids are specified by two or more codons. For example, both UUU and UUG stand
for the amino acid phenylalanine (Phe). Notice that the codon AUG not only
stands for the amino acid methionine (Met) but also functions as a signal to
"start" translating the RNA at that place. Three of the 64 codons
function as "stop" signals. Anyone of these termination codons marks
the end of a genetic message.
As Figure 10.11 shows, 61 of the 64
triplets code for amino acids. The triplet AUG has a dual function: It not only
codes for the amino acid methionine (Met) but can also provide a signal for the
start of a polypeptide chain. Three of the other codons do not designate amino
acids. They are the stop codons that instruct the ribosomes to end the
polypeptide.
In Figure 10.11 that there is redundancy
in the code but no ambiguity. For example, although codons UUU and UUC both
specify phenylananine (redundancy), neither of them ever represent any other
amino acid (no redundancy). The codons
in the figure are the triplets found in RNA. They have a straightforward,
complementary relationship to the codons in DNA. The nucleotides making up the
codons occur in a linear order along the DNA and RNA, with no gaps or
"punctuation" separating the codons.
Almost all of the genetic code is
shared by all organisms, from the simplest bacteria to the most complex
plants and animals. The universality of the genetic vocabulary suggests that it
arose very early in evolution and was passed on over the eons to all the
organisms living on Earth today. Such universality is extremely important to
modern DNA technologies. Because the code is the same in different species,
genes can be transcribed and translated after transfer from one species to
another, even when the organisms are as different as a bacterium and a human,
or a firefly and a tobacco plant (Figure 10.11.). This allows scientists to mix
and match genes from various species-a procedure with many useful applications.
Transcription:
From DNA to RNA
Let's look more closely at transcription,
the transfer of genetic information from DNA to RNA. An RNA molecule is
transcribed from a DNA template by a process that resembles the synthesis of a
DNA strand during DNA replication. Figure 10.13a is a close-up view of this
process. As with replication, the two DNA strands must first separate at the
place where the process will start. In transcription, however, only one of the
DNA strands serves as a template for the newly forming molecule. The
nucleotides that make up the new RNA molecule take their places one at a time
along the DNA template strand by forming hydrogen bonds with the nucleotide
bases there. Notice that the RNA nucleotides follow the same base-pairing rules
that govern DNA replication, except that U, rather than T, pairs with A. The
RNA nucleotides are linked by the transcription enzyme RNA polymerase.
Figure 10.13b is an overview of the
transcription of an entire gene. Special sequences of DNA nucleotides tell the
RNA polymerase where to start and where to stop the transcribing process.
Initiation of
Transcription The "start transcribing" signal
is a nucleotide sequence called a promoter, which is located in the DNA at the
beginning of the gene. A promoter is a specific place where RNA polymerase
attaches. The first phase of transcription, called initiation, is the
attachment of RNA polymerase to the promoter and the start of RNA synthesis.
For any gene, the promoter dictates which of the two DNA strands is to be
transcribed (the particular strand varying from gene to gene).
RNA Elongation
During the second phase of transcription, elongation, the RNA grows longer. As
RNA synthesis continues, the RNA strand peels away from its DNA template,
allowing the two separated DNA strands to come back together in the region
already transcribed.
Termination of
Transcription In the third phase, termination, the RNA
polymerase reaches a special sequence of bases in the DNA template called a
terminator. This sequence signals the end of the gene. At this point, the
polymerase molecule detaches from the RNA molecule and the gene.
In addition to producing RNA that encodes
amino acid sequences, transcription makes two other kinds of RNA that are
involved in building polypeptides. We discuss these kinds of RNA a little
later.
The
Processing of Eukaryotic RNA
In prokaryotic cells, which lack nuclei,
the RNA transcribed from a gene immediately functions as the messenger molecule
that is translated, called messenger RNA (mRNA). But this is not the case in
eukaryotic cells. The eukaryotic cell not only localizes transcription in the
nucleus but also modifies, or processes, the RNA transcripts there before
they move to the cytoplasm for translation by the ribosomes.
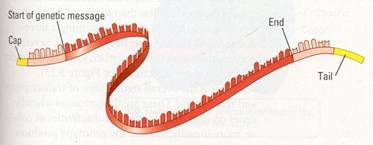
One kind of RNA processing is the addition
of extra nucleotides to the ends of the RNA transcript. These additions, called
the cap and tail, protect the RNA from attack by cellular enzymes and help
ribosomes recognize the RNA as mRNA.

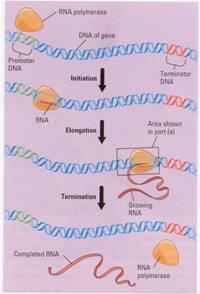
(a)
A close-up view of transcription (b)
Transcription of a gene
Figure 10.13 Transcription. (a) As ANA nucleotides base-pair one
by one with DNA bases on one DNA strand (called the template strand), the
enzyme ANA polymerase links the ANA nucleotides into an ANA chain. The orange
shape in the background is the ANA polymerase. (b) The transcription of an
entire gene occurs in r three phases: initiation, elongation, and termination
of the ANA. The section of DNA t where the ANA polymerase starts is called the
promoter; the place where it stops is , called the terminator.

Figure
10-14 Figure 10-15
Figure 10.14 The production of messenger RNA in a eukaryotic
cell. Both exons and introns are transcribed from the DNA. Additional
nucleotides, making up the cap and tail, are attached It the ends of the RNA
transcript. The exons are spliced together. The Iroduct, a molecule of
messenger RNA (mRNA), then travels to the :ytoplasm of the cell. There the
coding sequence will be translated.
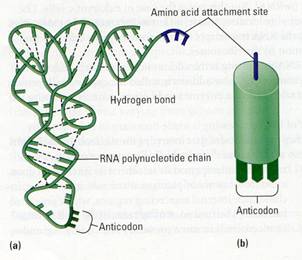
Figure 10.15 The structure of tRNA. (a) The RNA
polynucleotide is a "rope" whose appendages are the nitrogenous
bases. Dashed lines are hydrogen bonds, which connect some of the bases. The
site where an amino acid will attach is a three nucleotide segment at one end
(purple). Note the three-base anti codon at the bottom of the molecule (dark
green). The overall shape of a tRNA molecule is like the letter L. (b) This is
the representation of tRNA that we use in later diagrams.
Another type of RNA processing is made
necessary in eukaryotes by noncoding stretches of nucleotides that interrupt --
nucleotides that actually code for amino acids. It is as if unintelligible
sequences of letters were randomly interspersed in an otherwise intelligible
document. most genes of plants animals, It turns out, include such internal
noncoding regions, which are called introns. (The functions of introns,
if any, and how introns evolved remain a mystery.) The coding regions -- the
parts of a gene that are expressed - are called exons. As Figure 10.14
illustrates, both exons and introns are transcribed from DNA into RNA. However,
before the RNA leaves the nucleus, the introns are removed, and the exons are
joined to produce an mRNA molecule with a continuous coding sequence. This
process is called RNA splicing. RNA splicing is believed to playa significant
role in humans in allowing our approximately 35,000 genes to produce many
thousands more polypeptides. This is accomplished by varying the exons that are
included in the final mRNA. With capping, tailing, and splicing completed, the
"final draft" of eukaryotic mRNA is ready for translation.
Translation:
The Players
As we have already discussed, translation
is a conversion between different languages-from the nucleic acid language to
the protein language-and it involves more elaborate machinery than
transcription.
Messenger RNA (mRNA)
The first important ingredient required for translation is the mRNA produced by
transcription. Once it is present, the machinery I used to translate mRNA
requires enzymes and sources of chemical energy, such as ATP. In addition,
translation requires two heavy-duty components: ribosomes and a kind of RNA
called transfer RNA.
Transfer RNA (tRNA)
Translation of any language into another language requires an interpreter, person or device that can recognize the
words of one language and convert them into the other. Translation of the
genetic message carried in mRNA into the amino acid language of proteins also
requires an interpreter. To convert the three-letter words ( codons ) of
nucleic acids to the one-letter, amino acid words of proteins, a cell uses a
molecular interpreter, a type of RNA called transfer RNA. abbreviated tRNA
(Figure 10.15).
Figure
10-16 Figure
10-17
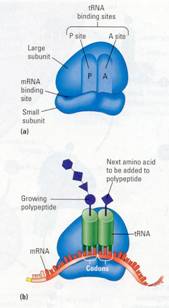
Figure 10.16 The ribosome. (a) A simplified diagram of a
ribosome, showing its two subunits and sites where mRNA and tRNA molecules
bind. (b) When functioning in polypeptide synthesis, a ribosome holds one
molecule of mRNA and two molecules of tRNA. The growing polypeptide is attached
to one of the tRNAs.
Figure 10.17 A molecule of mRNA. The pink ends are
nucleotides that are not part of the message; that is, they are not translated.
These nucleotides, along with the cap and tail (yellow),
help the mRNA attach to the ribosome.
A cell that is ready to have some of its
genetic information translated into polypeptides has in its cytoplasm a supply
of amino acids, either obtained from food or made from other chemicals. The
amino acids themselves cannot recognize the codons arranged in sequence along
messenger RNA. It is up to the cell's molecular interpreters, tRNA molecules,
to match amino acids to the appropriate codons to form the new polypeptide. To
perform this task, tRNA molecules must carry out two distinct functions: ( 1)
to pick up the appropriate amino acids, and (2) to recognize the appropriate
codons in the mRNA. The unique structure of tRNA molecules enables them to
perform both tasks-
As shown in Figure 10.15a, a tRNA molecule
is made of a single strand of RNA - one polynucleotide chain-consisting of
about 80 nucleotides. The chain twists and folds upon itself, forming several
double-stranded regions in which short stretches of RNA base-pair with other
stretches. At one end of the folded molecule is a special triplet of bases
called an anticodon. The anticodon triplet is complementary to a codon triplet
on mRNA. During translation, the anticodon on tRNA recognizes a particular
codon on mRNA by using base-pairing rules. At the other end of the tRNA
molecule is a site where an amino acid can attach. Although all tRNA molecules
are similar, there is a slightly different version of tRNA for each amino acid.
Ribosomes Ribosomes are the organelles that coordinate
the functioning of the mRNA and tRNA and actually make polypeptides. As you can
see in Figure 10.16a, a ribosome consists of two subunits. Each subunit is made
up of proteins and a considerable amount of yet another kind of RNA, ribosomal
RNA (rRNA). A fully assembled ribosome has a binding site for mRNA on its small
subunit and binding sites for tRNA on its large subunit. Figure 10.16b shows
how two tRNA molecules get together with an mRNA molecule on a ribosome. One of
the tRNA binding sites, the P site, holds the tRNA carrying the growing
polypeptide chain, while another, the A site, holds a tRNA carrying the next
amino acid to be added to the chain. The anticodon on each tRNA base pairs with
a codon on mRNA. The subunits of the ribosome act like a vise, holding the tRNA
and mRNA molecules close together. The ribosome can then connect the amino acid
from the A site tRNA to the growing polypeptide.
 ‘
‘
Figure
10.18 Figure 10.19
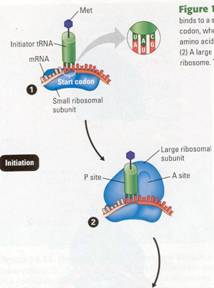
Figure 10.18 The initiation of translation. (1) An mRNA molecule
binds to a small ribosomal subunit. A special initiator tRNA then binds to the
start codon, where translation is to begin on the mRNA. The initiator tRNA
carries the amino acid methionine (Met); its anticodon, UAC, binds to the start
codon, AUG. (2) A large ribosomal
subunit binds to the small one, creating a functional ribosome. The initiator
tRNA fits into the P site on the ribosome.
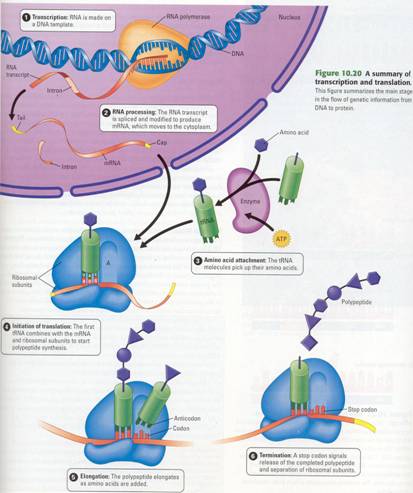
Figure
10.20
Translation:
The Process
Translation can be divided into the same
three phases as: transcription initiation, elongation, and termination.
Initiation
This first phase brings together the mRNA, the first amino acid with its
attached tRNA, and the two subunits of a ribosome. An mRNA molecule, even after
splicing, is longer than the genetic message it carries (Figure 10.17)
.Nucleotide sequences at either end of the molecule are not part of the
message, but along with the cap and tail in eukaryotes, they help the mRNA bind
to the ribosome. The initiation process determines exactly where translation will begin so that the mRNA codons will
be translated into the correct sequence of amino acids. Initiation occurs in
two steps, as shown in Figure 10.18.
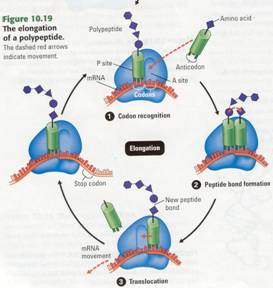
Elongation
Once initiation is complete, amino acids are added one by one to the first ammo
acid. Each addition occurs ill a three-step elongation process (Figure 10.19).
Step (1) Codon
recognition. The anticodon of an
incoming tRNA molecule, carrying its ammo acid, pairs with the mRNA codon in
the A site of the ribosome.
Step (2) Peptide bond formation. The polypeptide leaves the
tRNA in the P site and attaches to the amino acid on the tRNA in the A site.
The ribosome catalyzes bond formation. Now the chain has one more amino acid.
Step (3) Translocation. The P site tRNA now leaves the
ribosome, and the ribosome translocates (moves) the remaining tRNA, carrying
the growing polypeptide, to the P site. The mRNA and tRNA move as a unit. This
movement brings into the A site the next mRNA codon to be translated, and the
process can start again with step (1).
Termination
Elongation continues until a stop codon reaches the ribosome's A site. Stop.
codons-UAA, UAG, and UGA-do not code for amino acids but instead tell
translation to stop. The completed polypeptide, typically several hundred ammo
acids long, is freed, and the ribosome splits into its subunits.
Review:
DNA > RNA > Protein
Figure 10.20 reviews the flow of genetic
information in the cell, from DNA to RNA to protein. In eukaryotic cells,
transcription-the stage from DNA to RNA-occurs in the nucleus, and the RNA 8
Peptide bond formation is processed before it enters the cytoplasm. Translation
is rapid; a single ribosome can make an average-size polypeptide in less an a
minute. As it is made a polypeptide coils and folds, assuming a three-dimensional
shape, its tertiary structure. Several polypeptides may come together, forming
a protein with quaternary structure.
What is the overall significance of
transcription and transcription? These are the processes whereby genes control
the structures and activities of cells location or, more broadly, the way the
genotype produces the phenotype. The
chain command originates with information in the gene, a specific linear
sequence of nucleotides in the DNA. The
gene serves as a template, dictating the transcription of a complementary
sequence of nucleotides in mRNA. In turn, mRNA specifies the linear sequence in
which amino acids appear in a polypeptide. Finally, the proteins that form from
the polypeptides determine the appearance and capabilities of the cell and
organism.
Mutations
Since discovering how genes are translated
into proteins, scientists have been able to describe many heritable differences
in molecular terms. For instance, when a child is born with sickle-cell
disease, the condition can be traced back through a difference in a protein to
one tiny change in a gene. In one of the polypeptides in the hemoglobin
protein, the sickle-cell child has a single different amino acid. This
difference is caused by a single nucleotide difference in the coding strand of
DNA (Figure 10.21 ). In the double helix, a base pair is changed.
The sickle-cell allele is not a unique
case. We now know that the various alleles of many genes result from changes in
single base pairs in DNA. Any change in the nucleotide sequence of DNA is
called a mutation. Mutations can involve large regions of a chromosome or just
a single nucleotide pair, as in the sickle-cell allele.
Types of Mutations
Mutations within a gene can be divided into two general categories: base
substitutions and base insertions or deletions (Figure 10.22 ) .A base
substitution is the replacement of one base, or nucleotide, by another.
Depending on how a base substitution is translated, it can result in no change
in the protein, in an insignificant change, or in a change that might be
crucial to the life of the organism. Because of the redundancy of the genetic
code, some substitution mutations have no effect. For example, if a mutation
causes an mRNA codon to change from GAA to GAG, no change in the protein product
would result, because GAA and GAG both code for the same amino acid (Glu). Such
a change is called a silent mutation.
Other changes of a single nucleotide do
change the amino acid coding, Such mutations are called missense mutations. For
example, if a mutation causes an mRNA codon to change from GGC to AGC, the
resulting protein will have a serine (Ser) instead of a glycine (Gly) at this
position (see Figure 10.22a). Some missense mutations have little or no effect
on the resulting protein, but others, as we saw in the sickle-cell case, cause
changes in the protein that prevent it from performing normally.
Occasionally, a base substitution leads to
an improved protein or one with new capabilities that enhance the success of
the mutant organism and its descendants. Much more often, though, mutations are
harmful. Some base substitutions, called nonsense mutations, change an amino
acid codon into a stop codon. For example, if an AGA (Arg) codon is mutated to
a UGA ( stop) codon, the result will be a prematurely terminated protein, which
may not function properly.
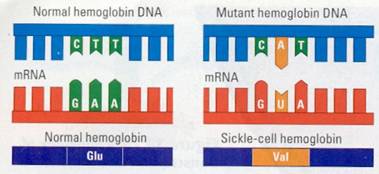

Figure 10.21 The molecular basis of sickle-cell disease. The
sickle-cell allele differs from its normal counter-part, a gene for hemoglobin,
by only one nucleotide, This difference changes the mRNA codon from one that
codes for the amino acid glutamic acid (Glu) to one that codes for valine
(Val).
Figure 10.22 Two types of mutations and their effects. Mutations
are changes in DNA, but they are represented here as reflected in mRNA and its
polypeptide product, (a)1n the base substitution shown here, an A replaces a G
in the fourth codon of the mRNA, The result in the polypeptide is a serine
(Ser) instead of a glycine (Gly), This amino acid substitution mayor may not
affect the protein's function, (b) When a nucleotide is deleted (or inserted),
the reading frame is altered, so that all the codons from that point on are
misread, The resulting polypeptide is likely to be completely nonfunctional,
Mutations involving the insertion or
deletion of one or more nucleotides in a gene often have disastrous effects.
Because mRNA is read as a series of nucleotide triplets during translation,
adding or subtracting nucleotides may alter the reading frame (triplet
grouping) of the genetic message. All the nucleotides that are
"downstream" of the insertion or deletion will be regrouped into
different codons. For example, consider an mRNA molecule containing the
sequence AAG-UUU-GGC-GCA; this codes for Lys-Phe-Gly-Ala. If a U is deleted in
the second codon, the resulting sequence will be AAG- UUG-GCG-CAU, which codes
for Lys-Leu-Ala-His (see Figure 10.22b). The altered polypeptide is likely to
be nonfunctional. Inserting one or two mRNA nucleotides would have a similarly
large effect.
Mutagens What
causes mutations? Mutagenesis, the creation of mutations, can occur in a number
of ways. Mutations resulting from errors during DNA replication or
recombination are known as spontaneous mutations, as are other mutations of
unknown cause. Other sources of mutation are physical and chemical agents
called mutagens. The most common physical mutagen is high-energy radiation,
such as X-rays and ultraviolet (UV) light. Chemical mutagens are of various
types. One type, for example, consists of chemicals that are similar to normal
DNA bases but that base-pair incorrectly when incorporated into DNA. Many
mutagens can act as carcinogens, agents that cause cancer. What can you do to
avoid exposure to mutagens? Several lifestyle practices can help, including
wearing protective clothing and sun screen to minimize direct exposure to the
sun’s rays and not smoking. But such precautions are not fool proof; for
example, you cannot entirely avoid UV radiation.
Although mutations are often harmful, they
are also extremely useful, both in nature and in the laboratory. Mutations are
the source of the rich diversity of genes in the living world, a diversity that
makes evolution by natural selection possible (Figure 10.23). Mutations are
also essential tools for geneticists. Whether naturally occurring or created in
the laboratory, mutations are responsible for the different alleles needed for
genetic research.